For the most part, X-ray crystallography has been the method of choice among scientists for determining the structure of proteins. This will cost a lot of money and take a lot of time. Naturally, the predictions made by AlphaFold would have to be validated using the tried-and-true methods.
Researchers have utilized AlphaFold, a revolutionary artificial intelligence (AI) network, to predict the structures of more than 200 million proteins from around 1 million species, representing nearly every known protein on Earth.
The data dump is freely accessible on a database created by DeepMind, the London-based artificial intelligence company that developed AlphaFold and is owned by Google, and the European Molecular Biology Laboratory’s European Bioinformatics Institute (EMBL–EBI), a non-profit organisation located near Cambridge, UK.
At a press briefing, DeepMind CEO Demis Hassabis stated, “Essentially, you can think of it as covering the entire protein universe.” The beginning of a new era of digital biology has arrived.
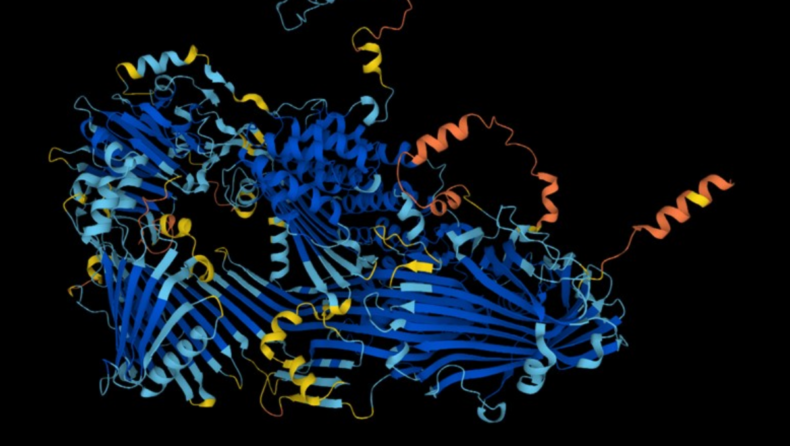
The cellular function of a protein is determined by its three-dimensional shape or structure. Most medications are produced based on structural information, and the construction of accurate maps of the amino-acid arrangement of proteins is frequently the first step in learning how proteins function.
DeepMind created the AlphaFold network using a form of artificial intelligence known as deep learning, and the AlphaFold database was released a year ago with more than 350,000 structure predictions covering nearly every protein produced by humans, mice, and 19 other organisms with extensive research.
The catalog now has approximately one million entries.
Christine Orengo, a computational biologist at University College London who has used the AlphaFold database to uncover novel protein families, says, “We’re preparing for the release of this massive trove.” It’s nice to have all the information forecasted for us.
Exemplary structures
Since the publication of AlphaFold a year ago, members of the life-sciences community have been hurrying to take advantage of the software. The network generates extremely precise predictions of the structures of several proteins.
It also gives information regarding the accuracy of its forecasts so that academics may determine whether they can be relied upon. X-ray crystallography and cryo-electron microscopy are time-consuming and expensive experimental approaches that have been traditionally used by scientists to determine protein structures.
According to EMBL–EBI, approximately 35 percent of the more than 214 million predictions are considered to be highly accurate, meaning that they are comparable to empirically established structures. Another 45% are deemed precise enough for numerous applications.